DNA synthesis

DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucleotide units are joined to form DNA; this can occur artificially (in vitro) or naturally (in vivo). Nucleotide units are made up of a nitrogenous base (cytosine, guanine, adenine or thymine), pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing (adenine and thymine, guanine and cytosine) occurs naturally when hydrogen bonds form between the nucleotide bases.
There are several different definitions for DNA synthesis: it can refer to DNA replication - DNA biosynthesis (in vivo DNA amplification), polymerase chain reaction - enzymatic DNA synthesis (in vitro DNA amplification) or gene synthesis - physically creating artificial gene sequences. Though each type of synthesis is very different, they do share some features. Nucleotides that have been joined to form polynucleotides can act as a DNA template for one form of DNA synthesis - PCR - to occur. DNA replication also works by using a DNA template, the DNA double helix unwinds during replication, exposing unpaired bases for new nucleotides to hydrogen bond to. Gene synthesis, however, does not require a DNA template and genes are assembled de novo.
DNA synthesis occurs in all eukaryotes and prokaryotes, as well as some viruses. The accurate synthesis of DNA is important in order to avoid mutations to DNA. In humans, mutations could lead to diseases such as cancer so DNA synthesis, and the machinery involved in vivo, has been studied extensively throughout the decades. In the future these studies may be used to develop technologies involving DNA synthesis, to be used in data storage.
DNA replication
[edit]

In nature, DNA molecules are synthesised by all living cells through the process of DNA replication. This typically occurs as a part of cell division. DNA replication occurs so, during cell division, each daughter cell contains an accurate copy of the genetic material of the cell. In vivo DNA synthesis (DNA replication) is dependent on a complex set of enzymes which have evolved to act during the S phase of the cell cycle, in a concerted fashion. In both eukaryotes and prokaryotes, DNA replication occurs when specific topoisomerases, helicases and gyrases (replication initiator proteins) uncoil the double-stranded DNA, exposing the nitrogenous bases.[1] These enzymes, along with accessory proteins, form a macromolecular machine which ensures accurate duplication of DNA sequences. Complementary base pairing takes place, forming a new double-stranded DNA molecule. This is known as semi-conservative replication since one strand of the new DNA molecule is from the 'parent' strand.
Continuously, eukaryotic enzymes encounter DNA damage which can perturb DNA replication. This damage is in the form of DNA lesions that arise spontaneously or due to DNA damaging agents. DNA replication machinery is therefore highly controlled in order to prevent collapse when encountering damage.[2] Control of the DNA replication system ensures that the genome is replicated only once per cycle; over-replication induces DNA damage. Deregulation of DNA replication is a key factor in genomic instability during cancer development.[3]
This highlights the specificity of DNA synthesis machinery in vivo. Various means exist to artificially stimulate the replication of naturally occurring DNA, or to create artificial gene sequences. However, DNA synthesis in vitro can be a very error-prone process.
DNA repair synthesis
[edit]Damaged DNA is subject to repair by several different enzymatic repair processes, where each individual process is specialized to repair particular types of damage. The DNA of humans is subject to damage from multiple natural sources and insufficient repair is associated with disease and premature aging.[4] Most DNA repair processes form single-strand gaps in DNA during an intermediate stage of the repair, and these gaps are filled in by repair synthesis.[4] The specific repair processes that require gap filling by DNA synthesis include nucleotide excision repair, base excision repair, mismatch repair, homologous recombinational repair, non-homologous end joining and microhomology-mediated end joining.
Reverse Transcription
[edit]Reverse transcription is part of the replication cycle of particular virus families, including retroviruses. It involves copying RNA into double-stranded complementary DNA (cDNA), using reverse transcriptase enzymes. In retroviruses, viral RNA is inserted into a host cell nucleus. There, a viral reverse transcriptase enzyme adds DNA nucleotides onto the RNA sequence, generating cDNA that is inserted into the host cell genome by the enzyme integrase, encoding viral proteins.[5]
Polymerase chain reaction
[edit]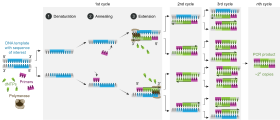
A polymerase chain reaction is a form of enzymatic DNA synthesis in the laboratory, using cycles of repeated heating and cooling of the reaction for DNA melting and enzymatic replication of the DNA.
DNA synthesis during PCR is very similar to living cells but has very specific reagents and conditions. During PCR, DNA is chemically extracted from host chaperone proteins then heated, causing thermal dissociation of the DNA strands. Two new cDNA strands are built from the original strand, these strands can be split again to act as the template for further PCR products. The original DNA is multiplied through many rounds of PCR.[1] More than a billion copies of the original DNA strand can be made.
Random mutagenesis
[edit]For many experiments, such as structural and evolutionary studies, scientists need to produce a large library of variants of a particular DNA sequence. Random mutagenesis takes place in vitro, when mutagenic replication with a low fidelity DNA polymerase is combined with selective PCR amplification to produce many copies of mutant DNA.[6]
RT-PCR
[edit]RT-PCR differs from conventional PCR as it synthesizes cDNA from mRNA, rather than template DNA. The technique couples a reverse transcription reaction with PCR-based amplification, as an RNA sequence acts as a template for the enzyme, reverse transcriptase. RT-PCR is often used to test gene expression in particular tissue or cell types at various developmental stages or to test for genetic disorders.[7]
Gene synthesis
[edit]Artificial gene synthesis is the process of synthesizing a gene in vitro without the need for initial template DNA samples. In 2010 J. Craig Venter and his team were the first to use entirely synthesized DNA to create a self-replicating microbe, dubbed Mycoplasma laboratorium.[8]
Oligonucleotide synthesis
[edit]Oligonucleotide synthesis is the chemical synthesis of sequences of nucleic acids. The majority of biological research and bioengineering involves synthetic DNA, which can include oligonucleotides, synthetic genes, or even chromosomes. Today, most synthetic DNA is custom-built using the phosphoramidite method by Marvin H. Caruthers. Oligos are synthesized from building blocks which replicate natural bases. Other techniques for snythesising DNA have been commercially made available, including Short Oligo Ligation Assembly.[9] The process has been automated since the late 1970s and can be used to form desired genetic sequences as well as for other uses in medicine and molecular biology. However, creating sequences chemically is impractical beyond 200-300 bases, and is an environmentally hazardous process. These oligos, of around 200 bases, can be connected using DNA assembly methods, creating larger DNA molecules.[10]
Some studies have explored the possibility of enzymatic synthesis using terminal deoxynucleotidyl transferase (TdT), a DNA polymerase that requires no template. However, this method is not yet as effective as chemical synthesis, and is not commercially available.[11]
With advances in artificial DNA synthesis, the possibility of DNA data storage is being explored. With its ultrahigh storage density and long-term stability, synthetic DNA is an interesting option to store large amounts of data. Although information can be retrieved very quickly from DNA through next generation sequencing technologies, de novo synthesis of DNA is a major bottleneck in the process. Only one nucleotide can be added per cycle, with each cycle taking seconds, so the overall synthesis is very time-consuming, as well as very error prone. However, if biotechnology improves, synthetic DNA could one day be used in data storage.[12]
Base pair synthesis
[edit]It has been reported that new nucleobase pairs can be synthesized, as well as A-T (adenine - thymine) and G-C (guanine - cytosine). Synthetic nucleotides can be used to expand the genetic alphabet and allow specific modification of DNA sites. Even just a third base pair would expand the number of amino acids that can be encoded by DNA from the existing 20 amino acids to a possible 172.[8] Hachimoji DNA is built from eight nucleotide letters, forming four possible base pairs. It therefore doubles the information density of natural DNA. In studies, RNA has even been produced from hachimoji DNA. This technology could also be used to allow data storage in DNA.[13]
References
[edit]- ^ a b Pelt-Verkuil, Evan (2008). "A Brief Comparison Between in Vivo DNA Replication and in Vitro PCR Amplification". Principles and Technical Aspects of PCR Amplification (PDF). Rotterdam: Springer Netherlands. pp. 9–15. doi:10.1007/978-1-4020-6241-4_2. ISBN 978-1-4020-6240-7. S2CID 215257488.
- ^ Patel, Darshil R.; Weiss, Robert S. (2018). "A tough row to hoe: when replication forks encounter DNA damage". Biochem Soc Trans. 46 (6): 1643–1651. doi:10.1042/BST20180308. PMC 6487187. PMID 30514768.
- ^ Reusswig, Karl-Uwe; Pfander, Boris (2019). "Control of Eukaryotic DNA replication Initiation - Mechanisms to Ensure Smooth Transitions". Genes (Basel). 10 (2): 99. doi:10.3390/genes10020099. PMC 6409694. PMID 30700044.
- ^ a b Tiwari V, Wilson DM 3rd. DNA Damage and Associated DNA Repair Defects in Disease and Premature Aging. Am J Hum Genet. 2019 Aug 1;105(2):237-257. doi: 10.1016/j.ajhg.2019.06.005. Review. PMID 31374202
- ^ Hughes, Stephen H (2015). "Reverse Transcription of Retroviruses and LTR Retrotransposons". Microbiology Spectrum. 3 (2): 1051–1077. doi:10.1128/microbiolspec.MDNA3-0027-2014. ISBN 9781555819200. PMC 6775776. PMID 26104704.
- ^ Forloni, M (2018). "Random Mutagenesis Using Error-prone DNA Polymerases". Cold Spring Harbor Protocols. 2018 (3): pdb.prot097741. doi:10.1101/pdb.prot097741. PMID 29496818.
- ^ Bachman, Julia (2013). Chapter Two - Reverse-Transcription PCR (RT-PCR). Methods in Enzymology. Vol. 530. pp. 67–74. doi:10.1016/B978-0-12-420037-1.00002-6. PMID 24034314.
- ^ a b Fikes, Bradley J. (May 8, 2014). "Life engineered with expanded genetic code". San Diego Union Tribune. Archived from the original on 9 May 2014. Retrieved 8 May 2014.
- ^ https://telesisbio.com/gibson-sola-platform/
- ^ Palluk, Sebastian; Arlow, Daniel H; et al. (2018). "De novo DNA synthesis using polymerase-nucleotide conjugates". Nature Biotechnology. 36 (7): 645–650. doi:10.1038/nbt.4173. OSTI 1461176. PMID 29912208. S2CID 49271982.
- ^ Perkel, Jeffrey M. (2019). "The race for enzymatic DNA synthesis heats up". Nature. 566 (7745): 565. Bibcode:2019Natur.566..565P. doi:10.1038/d41586-019-00682-0. PMID 30804572.
- ^ Tabatabaei, S. Kasra (2020). "DNA punch cards for storing data on native DNA sequences via enzymatic nicking". Nature Communications. 11 (1): 1742. Bibcode:2020NatCo..11.1742T. doi:10.1038/s41467-020-15588-z. PMC 7142088. PMID 32269230.
- ^ Hoshika, Shuichi (2020). "Hachimoji DNA and RNA. A Genetic System with Eight Building Blocks". Science. 363 (6429): 884–887. doi:10.1126/science.aat0971. PMC 6413494. PMID 30792304.
